Introduction
Surface reconstruction tool for 3DTK, is a program within 3DTK for reconstruction of triangulated mesh surfaces from 3D points data, with screened poisson surface reconstruction algorithm. It uses data structures and other tools that are provided with 3DTK. Users can use the recon program with the description in this page to reconstruct mesh surface from points. They can also include the poisson class like the recon program did in their own project to reach these functions.
Poisson surface reconstruction was introduced by Michael Kazhdan et. al. in 2006, and was improved by their team several times after that. Michael Kazhdan’s implementation of this algorithms has been widely used by many other software and tools like MeshLab and PCL, and is still under maintainance by now. The reconstruction algorithm of my tool is also based on it.
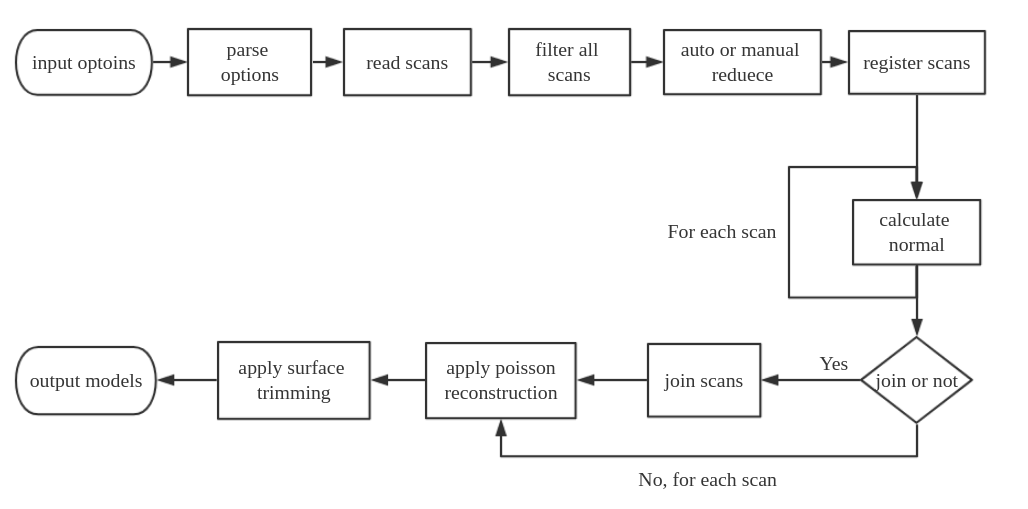
The general workflow of this tool is illustrated below. The programs start with users input options, it parses these options and set specified values or default values for all variables. Then it read scans, if user set range filter or reduction parameters, the program will do filter and reduction to get a smaller data size and range. After this, all scans will be registered based on frames or pose info. Then, normal calculation will be performed on each scan, if user chooses to join scans, all scans will be merged into a big dataset as the input of poisson reconstruction, if no join is set, each scan will be used as input of poisson reconstruction individually. After reconstruction, a surface trimming will be applied on the reconstructed models, and models will be written to a specific path in the end.
Sample usages
To use surface reconstruction tool in 3DTK, you can first download code with svn checkout https://svn.code.sf.net/p/slam6d/code/trunk slam6d-code and build the entire project with instruction in INSTALL.md.
Please know that this tool is not compatible with Ubuntu 14.04 (Trusty) due to a GCC compiler bug. If you are using Ubuntu 14.04, you may set CMake option WITH_MESH to OFF and try other functions of 3DTK.
- Use the default sample data under
dat/directory. This sample data is composed of three scans, we join them first with-jtag, since it is relatively sparse and non-closed, we set a trimming value 7 with-Ttag to trim extra part away. You can compare the model_all.obj and model_all_trimmed.obj to view the effects of trimming.bin/recon dat dat/test/model -d 12 -T 7.0 -j
- Use a common points data, for example the Stanford Bunny. A recommended way is convert the original model file into .xyz file that only contains the xyz coordinates, rename it as
scan000.3dand put it into a directory for exampledat/bunny/, new a file calledscan000.poseand put it under the same directory, and then use commands below. Unlike most scan data 3DTK is dealing with, the bunny surface is water-tight (closed), so we set the normal to be outward by adding a-Itag. For most scans data we use the default normal that are towards the scanner which we called inward.bin/recon dat/bunny dat/bunny/model -I
- Use Thermobremen data (No.22 on this page), a typical city scan data. This dataset is with file format uosr, we set it with
-ftag. Since it is relatively large, we set an auto reduction with-atag. We also set scans joining , poisson octree depth 12 and trimming value 9.0.bin/recon dat/testdata/thermobremen dat/testdata/thermobremen/result -f uosr -a -j -d 12 -T 9
- Use Thermocolorlab data (No.20 on this page), a typical indoor colored scan data. This dataset is with file format uos_rrgbt, so we set it with
-ffirst. This scan range is extremly large, we are only going to reconstruct surface within one room here, so we filter part of it with-utag, the filter range is -500 to 500 on x,y and z directions. We set scan number start from 0 and end with 12 with tag-sand-e, because other scans are not within the room we are going to show, so we just skip them for better speed and lower memory usages. We also set scan joing, octree depth and trimming value. To generate surface with color info, we have to use-Ctag.bin/recon dat/testdata/thermocolorlab/ dat/testdata/thermocolorlab/model -j -f uos_rrgbt -d 12 -T 8 -s 0 -e 12 -u "11;6;-500;500;-500;500;-500;500" -C
- Use Ostia data, another indoor colored scan data. Very similar to the Thermocolorlab data, this sample demonstrate that you could use only part of the scan by removing extra scans files. We first set file format to uos_rgb and use color as input data. Then set poisson octree depth 12 and trimming value 9.0.
bin/recon dat/testdata/ostia dat/testdata/ostia -j -f uos_rgb -d 12 -T 9 -C
- Use Randersacker data (No.26 on this page), a typical natrual open scene data. For this kind of large dataset, certain kinds of reduction and filtering is needed before reconstruction. You can either use
--aoption like Thermobremen data above, or to manually set reduction parameters with-roption and-Ooptions. To set a range filter, you can use-utag.
But sometimes for large dataset, reduction process will take a large number of time, finding an appropriate reduction parameters may take more time, so it is recommended that you usebin/scan_redprogram to reduce and filter scans first, then use the new files as data source for reconstruction.
Usages below usescan_redto reduce scans first, then usereconprogram to generate mesh surface.
Scan data of objects like tree leaves are not exactly ‘surface’ but more like a ‘volume’, so the reconstruction of tree leaves might be noisy. This sample is more likely to be a demo.bin/scan_red -s 0 -e 3 -f uosr --reduction OCTREE --voxel 20 --octree 10 dat/testdata/randersacker/ bin/recon dat/testdata/randersacker/reduced dat/testdata/randersacker/reduced/model -j -d 12 -T 9 -u "11;6;-1000;1000;-2000;2000;-1000;1000"
Options list
I/O options
- input-dir (mandatory), used without tag, this option provides the directory of input scans.
e.g.dat/test - output-dir, used without tag, this option provides the output file name of models, models will be written to the input directory with names like model0_all.obj, model1_all.obj, … (not joined) or model_all.obj and model_all_trimmed.obj (joined). if this option is not set, models will be written to the root directory of the project.
e.g.dat/test/model - –start or -s, used to set the start index of scan to be read, index start from 0, option value should be an integer number. By default this value will be 0.
e.g.-s 0 - –end or -e, used to set the end index of scan to be read, index start from 0, option value should be an integer number. By default this value will be -1, this means all scans in current directory will be read.
e.g.-e 10 - –format or -f, specify format of input data, option value should be a string, if not set, the default format will be uos. Supported formats description could be found here.
e.g.-f uos_rgb - –scanserver or -S, set to use scanserver as input method, no option value needed. Basic scan will be used if this tag is not setted.
e.g.-S - –max or -M, set to neglegt all data points with a distance larger than a certain value, option value should be an int number.
e.g.-M 100 - –min or -m, set to neglegt all data points with a distance smaller than a certain value, option value should be an int number.
e.g.-m 10 - –incolor or -C, set to use color from points and generate colored mesh, no option value needed. By default no color will be used.
e.g.-C - –nonormal or -N, set to disable normal output into model file, no option value needed. By default normal will be output.
e.g.-N
Reduction and filter options
- –autored or -a, set to use automatic reduction to reduce scan data, no option value needed. By default no reduction will be performed. Since automatic reduction might not fit all cases, set reduction parameters directly is a more recommended way.
e.g.-a - –reduce or -r, set to turns on octree based reduction, option value should be a double number represents the voxel size. This option should be used together with
--octree or -Ooption. By default no octree based reduction will be performed.
e.g.-r 20 - –octree or -O, set to use randomized octree based reduction, option value should be a int number represents the number of points per voxel. This option should be used together with
--reduce or -roption. By default the value should be 1.
e.g.-O 10 - –customFilter or -u, set to filter the scan data within a certain range, option value should be a string represents the filter parameters or a file that contains the filter parameters. By default no filter will be performed.
e.g.-u "11;6;-2000;2000;-2000;2000;-2000;2000"
Scan related options
- –join or -j, set to join all scans before reduction, no options value needed. By default scans will be reconstructed individually.
e.g.-j - –scale or -y, set to apply a scaling factor to the original data, option value should be a double number. By default no scaling oprations will be performed.
e.g.-y - –trustpose or -t, set to use pose info instead of frame info for transformation, no option value needed. This option should be used when no frames info is provided with input scan data. By default the frames info will be used to perform transformation for each scan.
e.g.-t
Normal calculation options
- –normal or -g, set to specify normal calculation methods, option value should be a string that represent the method. Available methods so far are KNN, ADAPTIVE_KNN, AKNN, ADAPTIVE_AKNN, PANORAMA (OpenCV required) and PANORAMA_FAST (OpenCV required). By default the AKNN methods will be used for normal calculation.
e.g.-g KNN - –K1 or -k, set k value used in the nearest neighbor search, option value should be an integer number. By default this value is 20.
e.g.-k 10 - –K2 or -K, set the max value of k adaptation, option value should be an integer number. By default this value is 20.
e.g.-K 10 - –width or -w, set the width of panorama image used for PANORAMA / PANORAMA_FAST normals calculation, option value should be an integer number. By default this value is 3600.
e.g.-w 2400 - –height or -h, set the height of panorama image used for PANORAMA / PANORAMA_FAST normals calculation, option value should be an integer number. By default thsi value is 1000.
e.g.-h 800
Poisson options
- –outward or -I, set to let surface normal outward from the object surface, no option value needed. By default the normals are towards the scanner which we called inward. In most scans data, normals of points are supposed to be inward towards the scanner. For some water-tight data that is usually composed of many scans already, for example a sculpture, their ideal normal should be outward from the object surface.
e.g.-I - –depth or -d, set the maximum depth of the octree that will be used for surface reconstruction, the value should be an integer number. By default this value is 8. As the octree depth increases, more detail of the surface could be revealed, but more memory / time will be used.
e.g.-d 12 - –samples or -P, set the minimum number of sample points that should fall within an octree node as the octree construction is adapted to sampling density, the option value should be a integer number. By default the this value is 1. For noise-free samples, small values in the range [1.0 - 5.0] can be used. For more noisy samples, larger values in the range [15.0 - 20.0] may be needed to provide a smoother, noise-reduced, reconstruction.
e.g.-P 10 - –trim or -T, set the value for mesh trimming, the option value should be an floating number. By default the trimming value is 0 which means no face will be trimmed. Since the poisson surface reconstruction tends to generate a closed surface that interpolate many extra faces that are far away and dense, the trimming operation would remove this part of faces based on a thresholding density value. As the trimming value increases, more faces will be trimmed and the remaining faces are fewer and fewer. Typically trimming value from 6 to 9 work for most scans data.
e.g.-T 7.0
Commits
Commits can be viewed on Source Forge commits list. Most of my source codes and changes are within include/mesh and src/mesh directories.
- [r1893] mesh: minor bug fixing and add README.md
- [r1892] Disable build of mesh with Ubuntu trusty
- [r1891] mesh: fix bool_switch issue in parse_options
- [r1890] mesh: fix bugs for color reading and transformation
- [r1889] mesh: reorganize source code file structures, fix bug for default PointWeight unset
- [r1886] mesh: improve auto reduce option and memory usages
- [r1883] mesh: add automatic scan reduction before reconstruction, fix some bugs with real data.
- [r1880] mesh: improve scan reduction in recon
- [r1878] mesh: Add options for input/output color data and output normal data
- [r1877] Apply SurfaceTrimmer with color to recon program
- [r1875] mesh: apply PoissonRecn with color to recon program
- [r1874] mesh: Add PoissonRecon with color support
- [r1868] normals: add flipNormal function and its usages in calc_normals program; mesh: add flipNormal option for surface inward or outward selection
- [r1859] mesh: unify poisson related data structures and remove outdated codes
- [r1855] mesh: add surface trimmer to filter reconstructed mesh
- [r1851] mesh: try to rebuild
- [r1850] mesh: update poisson reconstruction with density calculation version
- [r1846] mesh: add model face filter based on its distance to the nearest point
- [r1842] mesh: remove cgal related codes to avoid building failures with cgal version problems
- [r1841] mesh: add some other testing approaches for normal estimation and surface reconstruction
- [r1839] Seperate recon program out of poisson, enable all normal calculation methods.
- [r1829] mesh: remove eigenlib from mesh dir, use system installed eigen instead
- [r1828] mesh: fix wrong face indices issue
- [r1827] mesh: add simple mesh smoothing based on vcglib, for testing purpose
- [r1825] mesh: let scans joining work before reconstruction
- [r1823] revert mischanging of CMakeLists
- [r1822] mesh: add options for joining scans before surface reconstruction (joining not finished)
- [r1820] mesh: apply poisson reconstruction to each scan individually
- [r1818] Add parameter options for poisson
- [r1817] Add parameters for normal estimation
- [r1814] mesh: remove outdated PoissonRecon dir
- [r1813] mesh: integrate scanio and calc_normals together with current poisson
- [r1811] mesh: add missing poisson.h
- [r1810] mesh: add sample scanio and calc_normals
- [r1795] Integrate KB’s poisson reconstruction into src/mesh and add two sample test data
- [r1793] Update CMakeLists.txt to involve subdir ‘src/mesh’ in
- [r1792] Add mesh input, output and redering with sample data
Problems & Future work
1. Normal calculation: The current normal calculation is indeed very useful and robust. But it would be better if it can have a better identity near edges area. This problem is identical for sparse data. For real data with relatively high points density, normal calculation near edges looks relatively okay.
2. Scan joining: Current scan joining load all scans and perform a transformation on each of them and reach a final registration scan in the end. Maybe joining specific scans with real relationships would be more useful. For example, among 100 scans, scan004 to 009 generates all faces of a building, then we should cluster these scans together and calculate normals for them, this would avoid edge problem in the normal calculation. To reach this, some tag might be needed for each scan, that cluster a large scene into several water-tight objects.
3. Mesh rendering: Currently, all rendering results in this page are screenshots from MeshLab. It would be great if a basic mesh renderer could be added into show or qtshow. This way users can quickly view the surface they are just reconstructed. Or to be more advanced, surface may be rendered together with point set.
4. Mesh processing: Originally I was going to add some mesh processing functions, but after all, only a surface trimmer is truly added and used. Maybe some mesh processing tools could be added in the future, with CGAL that is already introduced into 3DTK. But the priority of mesh processing might not be very high because I think 3DTK is originally designed for point data, surface mesh should be the output, and further processing would be on the next stage of work.
5. Other poisson options I did not include all parameters of Poisson reconstruction into my work, because many of them are for research design purpose that might not be very useful for common data, and have not been truly tested. It would be great to test all of the options and provide interfaces for those useful ones.
6. Ohter reconstruction methods Poisson reconstruction is a typical implicit reconstruction method, some other explicit ones might be introduced in the future. Since different data might fit different methods. Within the mesh/poisson directory there is a SSDRecon, since it shares similar data structures and workflow with PoissonRecon, it might be good to also introduce it into recon program.
References papers
Elseberg, J., Borrmann, D., Nüchter, A.: One Billion Points in the Cloud – An Octree for Efficient Processing of 3D Laser Scans. Isprs Journal of Photogrammetry & Remote Sensing, 2013, 76(1):76-88.</p>
Kazhdan, M., Bolitho, M., Hoppe, H.: Poisson Surface Reconstruction.: Proceedings of the Eurographics Symposium on Geometry Processing 2006. 2006:61-70.
Kazhdan, M., Hoppe, H.: Screened Poisson Surface Reconstruction. Acm Transactions on Graphics, 2013, 32(3):1-13.
Borrmann, D., Hess, R., Eck, D., Houshiar, H., Nüchter, A., Schilling, K.: Evaluation of Methods for Robotic Mapping of Cultural Heritage Sites.Proceedings of the 2th IFAC conference on Embedded Systems, Computer Intelligence and Telematics (CESCIT ‘15). p. 105–110.